Andrew Speidell, Ph.D.
I am a molecular and behavioral neuroscientist who is focused on the complex set of mechanisms which underly neurodegeneration, especially those which affect synaptic transmission. Currently, my work investigates the molecular pathogenesis of Huntington’s Disease, with a focus on the heavily disrupted epigenetic landscape in the early stages of disease progression. To do so, I utilize a variety of computational approaches, cell-free experiments, in vitro systems, and an array of mouse models to bridge the gap in understanding between (epi)genetic modifications and phenotypic outcomes.
Some exciting images from my current and previous work:
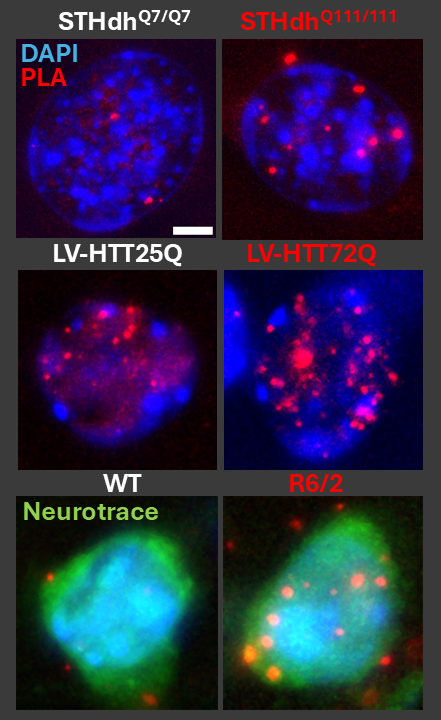
Figure 1: Visualizing intranuclear interactions of a neuronal DNA methyltransferase and a histone lysine N-methyltransferase in models of Huntington's disease. Each dot represents the proximity ligation assay product of two chromatin-remodeling enzymes within 50nm of one another, with a robust increase of this interaction in the indicated Huntington's disease models (red title) vs control (white title). (Top) STHdh striatal precursor-derived and differentiated cells. (Middle) Primary neurons transfected with 25- or 72- glutamine repeat HTTex1 lentiviral constructs. (Bottom) Neurons in the dorsolateral striatum of wildtype (WT) or R6/2 (HD model) mice.
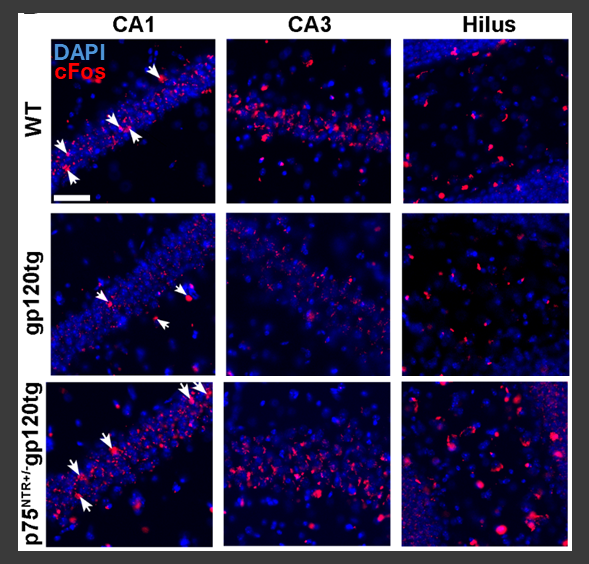
Figure 2: Reduced expression of the pan-proneurotrophin receptor p75NTR restores expression of the immediate early gene cFos in the hippocampus of gp120 transgenic mice during recall of an extinguished fear conditioning task. Expression of cFos is reduced in the hippocampus of gp120tg mice during recall of an extinguished association vs WT, but restored in mice lacking one P75NTR allele in the same epoch of the behavioral paradigm. See: Speidell et al 2024, J. Neurochemistry.
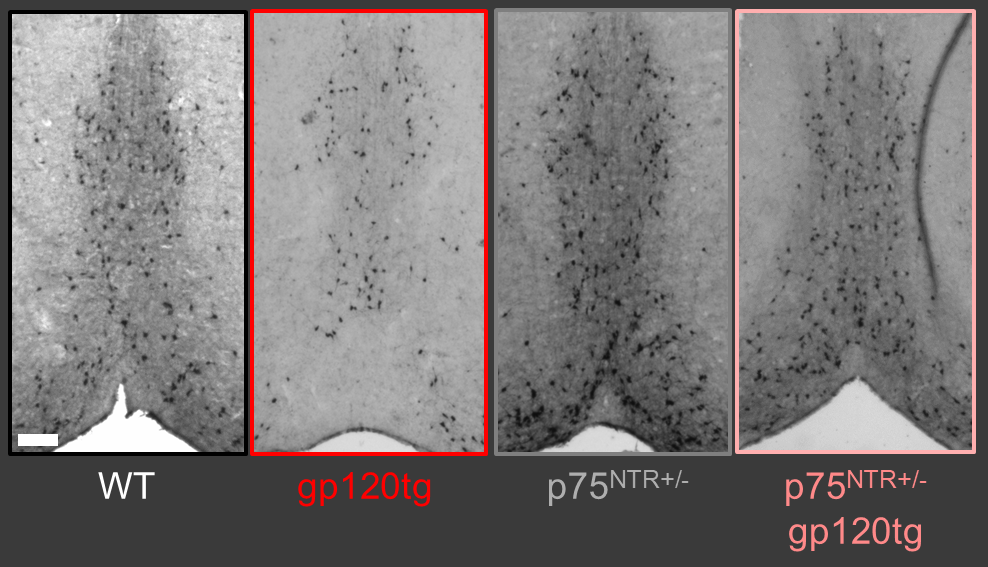
Figure 3: Widefield microscopy and stereological analysis of choline acetyltransferase-expressing cells of the basal forebrain indicates that this special neuronal population is sharply reduced in aged gp120tg mice, but their loss is sharply attenuated with reduction of p75NTR expression. Sections from aged mice. Bregma +1.0mm (A/P). See: Speidell et al 2024, J. Neurochemistry.
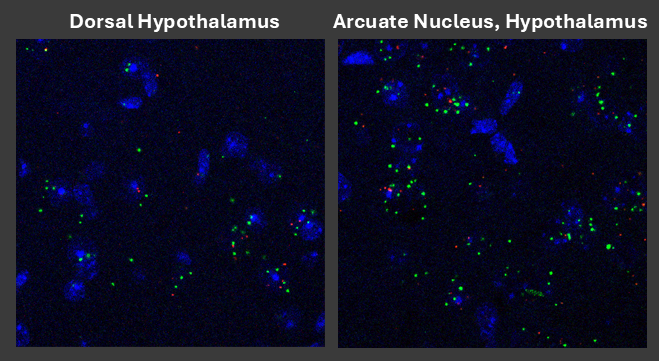
Figure 4: Confocal photomicrograph of the expresison and distribution of Gpr75 mRNA using RNAScope (TM) in young wild-type mice. Green puncta indicate Gpr75 mRNA, while red puncta indicate expression of the pan-neuronal marker mRNA Rbfox3. Expression of this transcript appears to be highly enriched in "feeding drive" centers of the hypothalamus.
